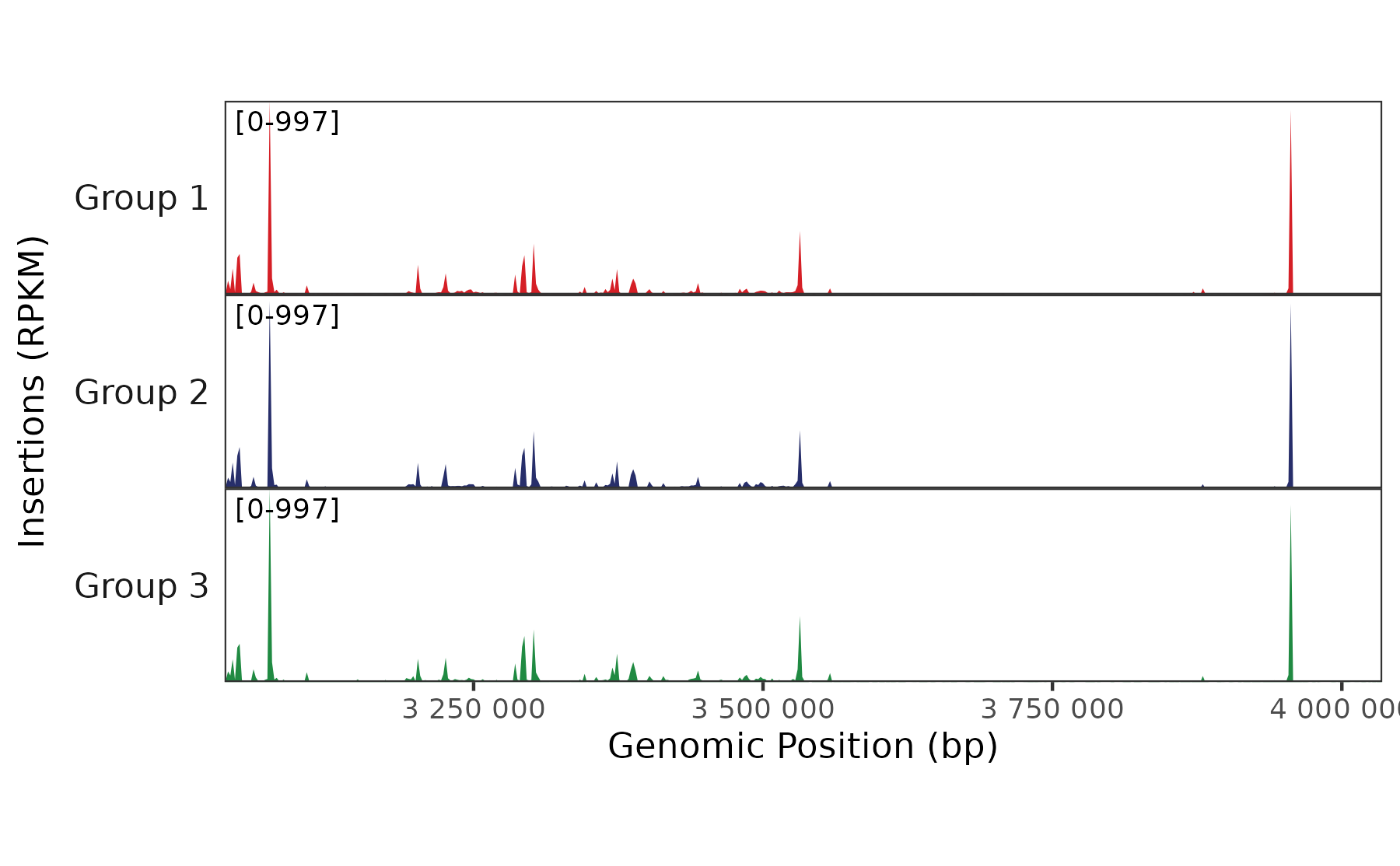
Plot a pseudobulk genome track, showing the number of fragment insertions across a region for each cell type or group.
Usage
trackplot_coverage(
fragments,
region,
groups,
cell_read_counts,
group_order = NULL,
bins = 500,
clip_quantile = 0.999,
colors = discrete_palette("stallion"),
legend_label = NULL,
zero_based_coords = !is(region, "GRanges"),
return_data = FALSE
)Arguments
- fragments
Fragments object
- region
Region to plot, e.g. output from
gene_region(). String of format "chr1:100-200", or list/data.frame/GRanges of length 1 specifying chr, start, end. Seehelp("genomic-ranges-like")for details- groups
Vector with one entry per cell, specifying the cell's group
- cell_read_counts
Numeric vector of read counts for each cell (used for normalization)
- group_order
Optional vector listing ordering of groups
- bins
Number of bins to plot across the region
- clip_quantile
(optional) Quantile of values for clipping y-axis limits. Default of 0.999 will crop out just the most extreme outliers across the region. NULL to disable clipping
- colors
Character vector of color values (optionally named by group)
- legend_label
Custom label to put on the legend (no longer used as color legend is not shown anymore)
- zero_based_coords
Whether to convert the ranges from a 1-based end-inclusive coordinate system to a 0-based end-exclusive coordinate system. Defaults to true for GRanges and false for other formats (see this archived UCSC blogpost)
- return_data
If true, return data from just before plotting rather than a plot.
- scale_bar
Whether to include a scale bar in the top track (
TRUEorFALSE)
Value
Returns a combined plot of pseudobulk genome tracks. For compatability with
draw_trackplot_grid(), the extra attribute $patches$labels will be added to
specify the labels for each track. If return_data or return_plot_list is
TRUE, the return value will be modified accordingly.
Examples
frags <- get_demo_frags()
## Use genes and blacklist to determine proper number of reads per cell
genes <- read_gencode_transcripts(
file.path(tempdir(), "references"), release = "42",
annotation_set = "basic",
features = "transcript"
)
blacklist <- read_encode_blacklist(file.path(tempdir(), "references"), genome="hg38")
read_counts <- qc_scATAC(frags, genes, blacklist)$nFrags
region <- "chr4:3034877-4034877"
cell_types <- paste("Group", rep(1:3, length.out = length(cellNames(frags))))
BPCells:::render_plot_from_storage(
trackplot_coverage(frags, region, groups = cell_types, cell_read_counts = read_counts),
width = 6, height = 3
)